-
摘要:
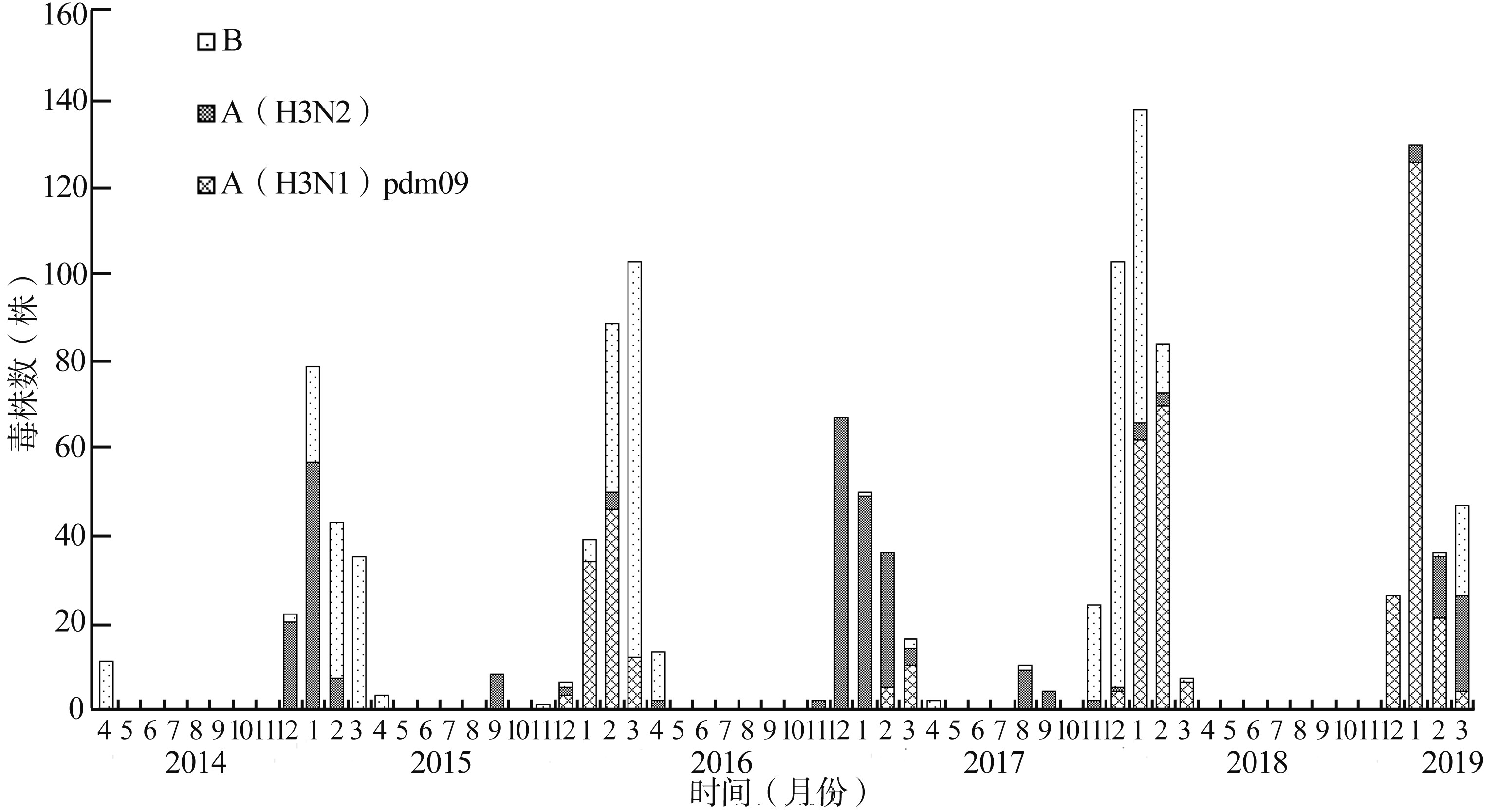
目的 了解2014-2019年青岛市人群A型流感病毒(influenza A virus, IAV)流行病学和遗传特征。 方法 提取9 807份流感样病例咽拭子标本的病毒RNA, 采用多重实时反转录PCR方法鉴定分型; 采用一步法反转录PCR扩增IAV血凝素(hemagglutinin, HA)和神经氨酸酶(neuraminidase, NA)基因, 并进行序列测定和基因分析。 结果 2014-2019年青岛市流感病毒流行具有北半球典型季节性流感特征。夏季小量流行主要是H3N2。青岛市IAV中H1N1pdm09和H3N2分别占57.6%和42.4%, 并呈现交替流行。基因群分属于6B和3C。两者HA和NA基因进化都处在净化选择下, 但抗原进化可能具有不同的进化模式。与2009年毒株相比, 2019年青岛市H1N1pdm09和H3N2的HA分别已发生21个和30个氨基酸替换, 均主要发生在头部, 分别包含5个和18个抗原位点。H1N1pdm09检测到两株神经氨酸酶抑制剂抗性突变, 1株为H275Y突变, 另1株为S247N突变; H3N2中未发现NAI抗性突变。 结论 H1N1pdm09和H3N2是2014-2019年青岛市季节性IAV中的主要组分, 进化迅速。加强流感监测和研究十分必要, 流感疫苗株需要及时更新。 Abstract:Objective To analyze the epidemiology and molecular characteristics of influenza A virus(IAV) circulated in Qingdao from 2014 to 2019. Methods Throat swab samples from 9 807 influenza-like cases were collected and viral RNA was extracted. All samples were screened for influenza virusby multiplex real-time reverse transcription PCR. Hemagglutinin(HA) and neuraminidase(NA) genes were amplifyed by one-step reverse transcription PCR and sequenced, and then molecular characteristics of them were analyzed. Results The characteristics of influenza virus in Qingdao from 2014 to 2019 were similar to those of the typical seasonal influenza in the northern hemisphere. A small peak of summer influenza was mainly attributed to H3 N2. Of these IAV positive cases, H1 N1 pdm09 and H3 N2 accounted for 57.6% and 42.4% respectively, both alternately popular in different years. Phylogenetic tree indicated that they belonged to the 6 B and 3 C subgroup respectively. The genetic evolution of H1 N1 pdm09 and H3 N2 was under purifying selection, but antigenic evolution might have different evolution patterns. Compared with strains in 2009, twenty-one and thirty amino acid substitutions in HA occurred respectively in H1 N1 pdm09 and H3 N2 viruses in 2019, and they were mainly located in the head domain, including 5 and 18 antigen sites respectively. Only one isolate with H275 Y mutation and one with S247 N known to reduce susceptibility to NA inhibitors(NAI) were detected in H1 N1 pdm09, and no NAI-resistant mutation was found in H3 N2. Conclusions H1 N1 pdm09 and H3 N2 are the most prevalent strains of seasonal IAV with rapid evolution in Qingdao. Strengthening influenza surveillance and research is necessary and influenza vaccine strains need to be updated in time. -
Key words:
- Influenza A virus /
- Hemagglutinin /
- Neuraminidase /
- Molecular evolution
-
表 1 2014-2019年青岛市流感病毒检测阳性率
Table 1. Positive rate of influenza virus in Qingdao from 2014 to 2019
年度 阳性数(例) 样本总数(例) 阳性率[%(95%CI)] 基因型(例) IBV pH1N1 H3N2 2014- 190 2 004 9.5(8.2~10.8) 106 0 84 2015- 249 2 026 12.3(10.9~13.7) 140 95 14 2016- 184 1 967 9.4(8.1~10.6) 14 15 155 2017- 372 1 890 19.7(17.9~21.5) 207 142 23 2018-2019 239 1 920 12.4(11.0~13.9) 22 177 40 合计 1 234 9 807 12.6(11.9~13.2) 489 429 316 表 2 2014-2019年青岛市pH1N1和H3N2基因进化速率和阳性选择分析
Table 2. Evolution rate and positive selection for HA and NA genes of pH1N1 and H3N2 viruses in Qingdao from 2014 to 2019
序列数 核苷酸替代速率(×10-3) 阳性选择位点 平均值 95%HPD SLAC FEL ω平均值 数量 位点 ω平均值 数量 位点 H1 60 3.56 2.61~4.46 0.240 0 - 0.240 0 - H3 54 2.12 1.55~2.69 0.242 0 - 0.242 0 - N1 60 2.80 2.07~3.56 0.368 0 - 0.368 0 - N2 54 2.51 1.82~3.24 0.175 0 - 0.175 0 - 表 3 pH1N1和H3N2基因群主要分支的HA特征位点变异
Table 3. Feature of amino acid substitutions identified in HAs of pH1N1 and H3N2 viruses
亚型 分支 特征变异位点 pH1N1 6B D97N、K163Q、S185T、S203T、A256T、K283E、E374K、S451N、E499K 6B.1 S84N、S162N、I216T 6B.2 V152T、V173I、E491G、D501E 6B.1A S74R、S164T、I295V H3N2 3C.3a T128A、A138S、R142G、N145S、F159S、N225D 3C.2a L3I、N145S、F159Y、K160T、N225D、Q311H、D489N 3C.2a1 N171K、I406V、G484E 3C.2a2 T131K、R142K、R261Q 3C.2a1b K92R、H311Q 注:D代表天冬氨酸, N代表天冬酰胺, K代表赖氨酸, Q代表谷氨酰胺, S代表丝氨酸, T代表苏氨酸, A代表丙氨酸, E代表谷氨酸, I代表异亮氨酸, V代表缬氨酸, G代表甘氨酸, R代表精氨酸, F代表苯丙氨酸, L代表亮氨酸, Y代表酪氨酸, Q代表谷氨酰胺, H代表组氨酸。 表 4 2019年青岛市pH1N1和H3N2毒株HA氨基酸位点变异
Table 4. Summary of amino acid substitutions identified in the HA of pH1N1 and H3N2 viruses in Qingdao in 2019
基因 毒株 氨基酸位点 H1 74 83 84 97 129 162 163 164 183 185 203 216 Cal09 S P S D N S K S S S S I 1QD2019 R S N N D/N N Q T P/S I/T T T 256 260 283 295 321 374 451 499 504 Cal09 A N K I I E S E K 1QD2019 T D/N E V V K N K R/K H3 3 33 45 48 62 92 121 128 131 142 144 145 Perth09 L Q S T K K N T T R K N 3QD2019 I R N I G R K T/A K/T G S S 159 160 171 183 194 197 198 212 214 219 223 225 Perth09 F K N L L Q A T S S V N 3QD2019 Y T K H L Q/R S A I S/F I D 278 311 312 347 406 484 489 529 Perth09 N Q N V I G D V 3QD2019 K Q S M/V V G/E N I/V 注:Cal09代表A/California/07/2009, 1QD2019代表2019年青岛pH1N1;Perth09代表A/Perth/16/2009, 3QD2019代表2019年青岛H3N2。D是天冬氨酸, N是天冬酰胺, K是赖氨酸, Q是谷氨酰胺, S是丝氨酸, T是苏氨酸, A是丙氨酸, E是谷氨酸, I是异亮氨酸, V是缬氨酸, G是甘氨酸, R是精氨酸, F是苯丙氨酸, L是亮氨酸, Y是酪氨酸, H是组氨酸, P是脯氨酸, M是甲硫氨酸。 -
[1] Segaloff H, Melidou A, Adlhoch C, et al. Co-circulation of inflenza A(H1N1)pdm09 andinflenza A(H3N2)viruses, world health organization(WHO)european region, October 2018 to February 2019[J]. Euro Surveill, 2019, 24(9): 1900125.DOI: 10.2807/1560-7917.ES.2019.24.9.1900125. [2] WHO Influenza Centre London. Report prepared for the WHO annual consultation on the composition of influenza vaccine for the northern hemisphere 2019-2020[EB/OL]. (2019-02-20)[2019-09-01]. https://www.crick.ac.uk/sites/default/files/2019-04/Crick%20VCMFeb2019%20report_toPost.pdf. [3] Mostafa A, Abdelwhab EM, Mettenleiter TC, et al. Zoonotic potential of influenza A viruses: acomprehensive overview[J]. Viruses, 2018, 10(9): 497.DOI: 10.3390/v10090497. [4] Petrova VN, Russell CA. The evolution of seasonal influenza viruses[J]. Nat Rev Microbiol, 2018, 16(1): 47-60.DOI: 10.1038/nrmicro.2017.118. [5] Hoffmann E, Stech J, Guan Y, et al. Universal primer set for the full-length amplification of all influenza A viruses[J]. Arch Virol, 2001, 146(12): 2275-2289.DOI: 10.1007/s007050170002. [6] Caini S, Huang QS, Ciblak MA, et al. Epidemiological and virological characteristics of influenza B: results of the global influenza B study[J]. Influenza and Other Respiratory Viruses, 2015, 9(1): 3-12.DOI: 10.1111/irv.12319. [7] Smith DJ, Lapedes AS, de Jong JC, et al. Mapping the antigenic and genetic evolution of influenza virus[J]. Science, 2004, 305(5682): 371-376.DOI: 10.1126/science.1097211. [8] Gamblin SJ, Haire LF, Russell RJ, et al. The structure and receptor binding properties of the 1918 influenza hemagglutinin[J]. Science, 2004, 303(5665): 1838-1842.DOI: 10.1126/science.1093155. [9] McAuley JL, Gilbertson BP, Trifkovic S, et al. Inflenza virus neuraminidase structure and functions[J]. Front Microbiol, 2019, 10: 39.DOI: 10.3389/fmicb.2019.00039. [10] Toledo-Rueda W, Rosas-Murrieta NH, Muǹoz-Medina JE, et al. Antiviral resistance markers in inflenza virussequences in Mexico, 2000–2017[J]. Infect Drug Resist, 2018, 11: 1751-1756.DOI: 10.2147/IDR.S153154. [11] Ye C, Zhu W, Yu J, et al. Understanding the complex seasonality of seasonal influenza A and B virus transmission: evidence from six years of surveillance data in Shanghai, China[J]. Int J Infect Dis, 2019, 81: 57-65.DOI: 10.1016/j.ijid.2019.01.027. [12] Lai JCC, Karunarathna HMTK, Wong HH, et al. Neuraminidase activity and specificity of influenza A virus are influenced by haemagglutinin-receptor binding[J]. Emerg Microbes Infect, 2019, 8(1): 327-338.DOI: 10.1080/22221751.2019.1581034. [13] Al Khatib HA, Al Thani AA, Gallouzi I, et al. Epidemiological and genetic characterization of pH1N1 and H3N2 influenza viruses circulated in MENA region during 2009–2017[J]. BMC Infect Dis, 2019, 19(1): 314.DOI: 10.1186/s12879-019-3930-6. [14] Li X, Chen J, Hu J, et al. Rapid evolution and gene communication of H3N2 and H1N1 influenza A viruses[J]. J Infect, 2019, 78(6): 491-503.DOI: 10.1016/j.jinf.2019.03.009. [15] Bedford T, Suchard MA, Lemey P, et al. Integrating influenza antigenic dynamics with molecular evolution[J]. eLife, 2014, 3: e01914.DOI: 10.7554/eLife.01914. [16] Bedford T, Riley S, Barr IG, et al. Global circulation patterns of seasonal influenza viruses vary with antigenic drift[J]. Nature, 2015, 523(7559): 217–220.DOI: 10.1038/nature14460. [17] Wei CJ, Boyington JC, Dai K, et al. Cross-neutralization of 1918 and 2009 influenza viruses: role of glycans in viral evolution and vaccine design[J]. Sci Transl Med, 2010, 2(24): 24ra21.DOI: 10.1126/scitranslmed.3000799. [18] Koel BF, Burke DF, Bestebroer TM, et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution[J]. Science, 2013, 342(6161): 976-979.DOI: 10.1126/science.1244730. [19] Luksza M, Lassig MA. Predictive fitness model for influenza[J]. Nature, 2014, 507(7490): 57-61.DOI: 10.1038/nature13087. [20] Liu Y, Childs RA, Matrosovich T, et al. Altered receptor specificity and cell tropism of D222 Ghemagglutinin mutants isolated from fatal cases of pandemic a(H1N1)2009 influenza virus[J]. J Virol, 2010, 84(22): 12069-12074.DOI: 10.1128/JVI.01639-10. [21] Trucchi C, Paganino C, Amicizia D, et al. Universal influenza virus vaccines: what needs tohappen next[J]. Expert Opin Biol Ther, 2019, 19(7): 671-683.DOI: 10.1080/14712598.2019.1604671. [22] Hua S, Li X, Liu M, et al. Antigenic variation of the human influenza A(H3N2)virus during the 2014-2015 winter season[J]. Sci China Life Sci, 2015, 58(9): 882-888.DOI: 10.1007/s11427-015-4899-z. -





 下载:
下载: