Epidemiological and genetic characteristics of norovirus in sentinel surveillance cases of viral diarrhea in Hubei Province from 2017 to 2019
-
摘要:
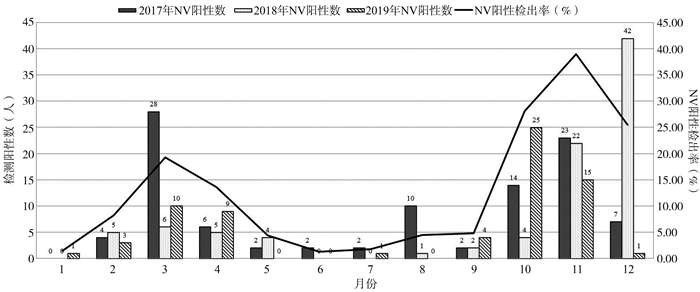
目的 了解湖北省2017-2019年病毒性腹泻哨点监测病例中诺如病毒(norovirus, NV)的流行病学特征和基因型别特征。 方法 收集2017年1月1日-2019年12月30日成人和儿童腹泻患者的粪便标本共1 957份,采用实时荧光RT-PCR法进行NV GI和GII型别鉴定;对NV检测阳性的GII型标本应用RT-PCR扩增开放阅读框(open reading frame, ORF)1/ORF2铰链区,将获得的PCR产物直接测序并进行基因型别和重组分析。 结果 男性和女性腹泻患者中NV阳性检出率差异有统计学意义(χ2 =5.18, P=0.023)。各年龄组腹泻患者间NV阳性检出率差异有统计学意义(χ2 =98.04, P<0.001),其中1~<2岁年龄组腹泻患者NV阳性检出率最高,为23.50%。2017-2019年湖北省NV感染呈现2个流行峰,高峰出现在10-12月间,低峰出现在3-4月。NV主要为GII基因型,其中以GII.4、GII.2和GII.3亚型为主。主要的重组型有GII.2[P16]、GII.4[P31]、GII.3[P12]和GII.4[P16]。 结论 2017-2019年湖北地区存在多种基因型NV的感染,并发现不同的重组亚型。加强NV的基因型别监测和重组分析有利于掌握NV的分子流行病学特征,为科学防控提供理论依据。 Abstract:Objective To describe the epidemiologic and genetic characteristics of norovirus (NV) in sentinel surveillance cases of viral diarrhea in Hubei Province from 2017 to 2019. Methods A total of 1 957 fecal samples from adult and child patients with diarrhea were collected from January 1, 2017 to December 30, 2019. Samples were detected using real-time RT-PCR. Positive samples of NV GII were amplified to obtain the sequences of open reading frame (ORF)1/ORF2 junction by RT-PCR. The obtained PCR products were directly sequenced and analyzed for genotype and recombination. Results There was a statistically significant difference in the detection rate of NV between males and females with diarrhea (χ2 =5.18, P=0.023). There is a statistically significant difference in the detection rate of NV between patients with diarrhea in each age group (χ2 =98.04, P < 0.001), among them, the 1~ < 2 year old age group has the highest NV-positive detection rate (23.50%) in patients with diarrhea. From 2017 to 2019, there were two epidemic peaks of NV infection in Hubei Province, the peak appeared in October-December, and the low peak appeared in March-April. The main NV genotypes were GII genogroup, of which GII.4, GII.2 and GII.3 were the main subtypes. The main recombination types were GII.2[P16], GII.4[P31], GII.3[P12] and GII.4[P16]. Conclusions There were multiple genotypes of NV infection in Hubei province from 2017 to 2019 and different recombination subtypes were found. Enhancing the monitoring of genotype and recombination of NV contributes to know the molecular epidemiology features of NV and provide theoretical basis for scientific prevention and control. -
Key words:
- Norovirus /
- Epidemiology /
- Genotype /
- Gene recombination
-
表 1 2017-2019年湖北省各年龄组腹泻患者粪便标本中NV检出情况
Table 1. Detection results of NV in fecal samples from patients with diarrhea in different age groups in Hubei Province from 2017 to 2019
年龄组(岁) 腹泻患者数(人) NV检测阳性数(人) NV阳性检出率(%) 0~<6月龄 221 8 3.62 6月龄~<1 173 16 9.25 1~<2 366 86 23.50 2~<5 408 63 15.44 5~<14 255 56 21.96 14~<20 51 4 7.84 20~<50 247 15 6.07 ≥50 236 12 5.08 合计 1 957 260 13.29 表 2 2017-2019年湖北省腹泻患者NV各月检测结果
Table 2. Monthly test results of NV in diarrhea patients in Hubei Province from 2017 to 2019
月份(月) 腹泻患者数 (人) NV检测阳性数 (人) NV阳性检出率 (%) 1 70 1 1.43 2 145 12 8.28 3 228 44 19.30 4 147 20 13.61 5 135 6 4.44 6 153 2 1.31 7 164 3 1.83 8 247 11 4.45 9 165 8 4.85 10 153 43 28.10 11 154 60 38.96 12 196 50 25.51 合计 1 957 260 13.29 -
[1] Patel MM, Widdowson MA, Glass RI, et al. Systematic literature review of role of noroviruses in sporadic gastroenteritis[J]. Emerg Infect Dis, 2008, 14(8): 1224-1231. DOI: 10.3201/eid1408.071114. [2] Robilotti E, Deresinski S, Pinsky BA. Norovirus[J]. Clin Microbiol Rev, 2015, 28(1): 134-164. DOI: 10.1128/CMR.00075-14. [3] Mans J. Norovirus infections and disease in lower-middle and low-income countries, 1997-2018[J]. Viruses, 2019, 11(4): 341. DOI: 10.3390/v11040341. [4] Xi JN, Graham DY, Wang KN, et al. Norwalk virus genome cloning and characterization[J]. Science, 1990, 250(4987): 1580-1583. DOI: 10.1126/science.2177224. [5] Kobayashi M, Matsushima Y, Motoya T, et al. Molecular evolution of the capsid gene in human norovirus genogroup Ⅱ[J]. Sci Rep, 2016, 6: 29400. DOI: 10.1038/srep29400. [6] Bull RA, Hansman GS, Clancy LE, et al. Norovirus recombination in ORF1/ORF2 overlap[J]. Emerg Infect Dis, 2005, 11(7): 1079-1085. DOI: 10.3201/eid1107.041273. [7] Chhabra P, de Graaf M, Parra GI, et al. Updated classification of norovirus genogroups and genotypes[J]. J Gen Virol, 2019, 100(10): 1393-1406. DOI: 10.1099/jgv.0.001318. [8] Rooney BL, Pettipas J, Grudeski E, et al. Detection of circulating norovirus genotypes: hitting a moving target[J]. Virol J, 2014, 11: 129. DOI: 10.1186/1743-422X-11-129. [9] Cannon JL, Barclay L, Collins NR, et al. Genetic and epidemiologic trends of norovirus outbreaks in the United States from 2013 to 2016 demonstrated emergence of NVel GⅡ. 4 recombinant viruses[J]. J Clin Microbiol, 2017, 55(7): 2208-2221. DOI: 10.1128/JCM.00455-17. [10] Liu L, Oza S, Hogan D, et al. Global, regional, and national causes of under-5 mortality in 2000-15: an updated systematic analysis with implications for the Sustainable Development Goals[J]. Lancet, 2016, 388(10063): 3027-3035. DOI: 10.1016/S0140-6736(16)31593-8. [11] Bull RA, Tanaka MM, White PA. Norovirus recombination[J]. J Gen Virol, 2007, 88(12): 3347-3359. DOI: 10.1099/vir.0.83321-0. [12] White PA. Evolution of norovirus[J]. Clin Microbiol Infect, 2014, 20(8): 741-745. DOI: 10.1111/1469-0691.12746. [13] Arana A, Cilla G, Montes M, et al. Genotypes, recombinant forms, and variants of norovirus GⅡ. 4 in Gipuzkoa (Basque Country, Spain), 2009-2012[J]. PLoS One, 2014, 9(6): e98875. DOI: 10.1371/journal.pone.0098875. [14] Chhabra P, Walimbe AM, Chitambar SD. Molecular characterization of three NVel intergenotype norovirus GⅡ recombinant strains from western India[J]. Virus Res, 2010, 147(2): 242-246. DOI: 10.1016/j.virusres.2009.11.007. [15] 白爱丽. 天津市北辰区一起学校诺如病毒感染疫情分析[J]. 河南预防医学杂志, 2020, 31(10): 784-785. DOI: 10.13515/j.cnki.hnjpm.1006-8414.2020.10.023.Bai AL. Analysis of norovirus infection in a school in Beichen District, Tianjin[J]. Henan J Prevent Med, 2020, 31(10): 784-785. DOI: 10.13515/j.cnki.hnjpm.1006-8414.2020.10.023. [16] 刘昊晖, 陈婷, 覃健敏, 等. 2018年南宁市诺如病毒聚集性疫情和暴发疫情流行特征分析[J]. 职业与健康, 2020, 36(14): 1987-1990. DOI: 10.13329/j.cnki.zyyjk.2020.0531.Liu HH, Chen T, Qin JM, et al. Analysis of epidemic characteristics of norovirus clusters and outbreaks in Nanning City in 2018[J]. Occupation Health, 2020, 36(14): 1987-1990. DOI: 10.13329/j.cnki.zyyjk.2020.0531. [17] 曹亿会, 杨景晖, 姜黎黎, 等. 2017-2019年我国西南地区病毒性腹泻病原检测分析[J]. 公共卫生与预防医学, 2021, 32(1): 10-13. https://www.cnki.com.cn/Article/CJFDTOTAL-FBYF202101003.htmCao YH, Yang JH, Jiang LL, et al. Analysis of pathogen detection of viral diarrhea in Southwest China from 2017 to 2019[J]. J Pub Health and Prev Med, 2021, 32(1): 10-13. https://www.cnki.com.cn/Article/CJFDTOTAL-FBYF202101003.htm [18] 金文军, 彭华, 王瑞琴, 等. 2018年北京市昌平区诺如病毒感染聚集性疫情流行病学特征[J]. 职业与健康, 2020, 36(7): 913-916. https://www.cnki.com.cn/Article/CJFDTOTAL-ZYJK202007016.htmJin WJ, Peng H, Wang RQ, et al. Epidemiological characteristics of clustered epidemics of norovirus infection in Changping District of Beijing in 2018[J]. Occupation Health, 2020, 36(7): 913-916. https://www.cnki.com.cn/Article/CJFDTOTAL-ZYJK202007016.htm [19] 李继耀, 刘蕴博, 赵翊祺, 等. 沈阳地区2015年-2017年诺如病毒感染性腹泻监测结果分析[J]. 中国卫生检验杂志, 2019, 29(15): 1884-1885, 1889. https://www.cnki.com.cn/Article/CJFDTOTAL-ZWJZ201915028.htmLi JY, Liu YB, Zhao YQ, et al. Monitoring results analysis of norovirus infected diarrhea in Shenyang from 2015 to 2017[J]. Chin J Health Lab Technol, 2019, 29(15): 1884-1885, 1889. https://www.cnki.com.cn/Article/CJFDTOTAL-ZWJZ201915028.htm [20] Lindesmith LC, Brewer-Jensen PD, Mallory ML, et al. Antigenic characterization of a novel recombinant GⅡ. P16-GⅡ. 4 Sydney norovirus strain with minor sequence variation leading to antibody escape[J]. J Infect Dis, 2018, 217(7): 1145-1152. DOI: 10.1093/infdis/jix651. [21] Barreira DMPG, Fumian TM, Tonini MAL, et al. Detection and molecular characterization of the novel recombinant norovirus GⅡ. P16-GⅡ. 4 Sydney in southeastern Brazil in 2016[J]. PloS one, 2017, 12(12): e189504. DOI: 10.1371/journal.pone.0189504. [22] Bidalot M, Théry L, Kaplon J, et al. Emergence of new recombinant noroviruses GⅡ. p16-GⅡ. 4 and GⅡ. p16-GⅡ. 2, France, winter 2016 to 2017[J]. Euro surveillance, 2017, 22(15): 30508. DOI: 10.2807/1560-7917.ES.2017.22.15.30508. [23] Medici MC, Tummolo F, Martella V, et al. Emergence of novel recombinant GⅡ. P16_GⅡ. 2 and GⅡ. P16_GⅡ. 4 Sydney 2012 norovirus strains in Italy, winter 2016/2017[J]. New Microbiol, 2018, 41(1): 71-72. http://www.newmicrobiologica.org/PUB/allegati_pdf/2018/1/71.pdf [24] Cheung SKC, Kwok K, Zhang LY, et al. Higher viral load of emerging norovirus GⅡ. P16-GⅡ. 2 than pandemic GⅡ. 4 and epidemic GⅡ. 17, Hong Kong, China[J]. Emerg Infect Dis, 2019, 25(1): 119-122. DOI: 10.3201/eid2501.180395. -





 下载:
下载: